Introduction
Overview
Teaching: 20 min
Exercises: 30 minQuestions
How do I start working in Galaxy?
What does my raw sequencing data looks like?
Do I have good enough quality and sequencing depth for my assembly?
Objectives
Getting familiar with Galaxy’s workflow and history
Understand the data generated and make a decision on how to process MinION data
Introduction
For the bioinformatics analysis of your genomes, we will be working in Galaxy.
There are two main online galaxy platforms; the global server: https://usegalaxy.org/ and the European server: https://usegalaxy.eu/. The global version has a slightly larger set of programs than the European version. However, the European version runs in general slightly faster, and therefore you are recommended to use the European server.
Overview of Genome Assembly Processes
Next-generation Sequencing (NGS) generates DNA reads in parallel, which are massive amount of short sequences of nucleotides. The length of these reads depends on the sequencing platforms as well as the methods used for extracting the DNA. Short-read NGS platform such as Illumina generate an average read length of 2 x 150 bp to 2 x 300 bp with high accuracy. In the past decade, long-read sequencing have become a reality, with the two leading platform are PacBio and Oxford Nanopore Technologies. Current PacBio HiFi can generate up to 25kb reads with high accuracy, while Nanopore can generate ultra long reads (>900 kb) but with less accuracy.
Long-read sequencing are crucial to solve assembly of complex genomic regions such as repeats, something that short-read could not achieve. A hybrid assembly (combining both the accuracy of short-reads and with long-reads to connect fragments) are used to generate a complete, high-quality genome. Nevertheless, the field is moving rapidly, opening a new avenues to look at microbial diversity at strain-level resolutions.
Because Nanopore sequencing results in so much longer reads, different bioinformatics tools need to be applied to process the data, as compared to the data generated by Illumina sequencing. Therefore, be careful to use bioinformatics programs, which are applicable for short or long reads depending on the dataset you have. In this course we are solely working with Nanopore sequencing.
More information on sequencing techniques:
Ryan Wick provides a nice “Guide to bacterial genome assembly” as a starting point to choose which approaches and techniques to assemble your genome. He also write an approach in combining ONT long reads with Illumina short reads to achieve perfect assemblies: Assembling the perfect bacterial genome using Oxford Nanopore and Illumina sequencing.
The general pipeline for processing nanopore reads into whole genomes are:
- Trimming and filtering reads
- Assembly of the reads into a genome
- Polishing the assembly
- Analysing the genome
We will be using similar pipeline to the one suggested by Ryan Wick with several measures to check quality in between each steps.
Importing Data into Galaxy
There are several ways we can input our data into Galaxy. We can upload datasets from our local machine, fetch from a public repository such as Zenodo, or reuse data from Galaxy history.
We will inform you how to fetch your raw sequencing data when it is ready. In the meantime, we can do an exercise using the data from previous year (2022), which are available from Zenodo. We will be using the strain with decent reads (barcode02).
Let’s login to Galaxy and import this data into our history:
- Go to https://usegalaxy.eu/
- Click
Login or Register. Create an account and activate (if you hadn’t so) - Import the data by pressing the
Upload Databutton on the upper right corner and choose thePaste/Fetch databutton at the bottom window. - Change the name from
New filetobarcode02.fastq.gzand paste the zenodo link:https://zenodo.org/record/6475902/files/barcode02.fastq.gzinto the box. - Press start to download
Discussion 01
Each item in the Zenodo repository are the sample raw reads that has been demultiplexed (one strain each).
- How many gigabytes of data are there?
- How much data was generated for an average sample?
- Why do we have uneven distribution of data for each samples?
- What kind of information can you get from a FASTQ file? What are the differences with a FASTA format?
Solution
TBD
- There are two main data formats which are relevant for your bioinformatics analysis. The first format is called FASTQ. It is usually used to store the raw reads (that is the nucleotide sequence string) together with their estimated quality(represented as ASCII bytes – not part of the curriculum). Data in fastq format can be identified by the endings: *.fq or *.fastq. The other format is called fasta. It is used to store any sort of DNA or amino acid sequence. Data in fasta format can be identified by the endings: *.fasta, *.fa, *.fna, *.fas or *.fsa. To save storage space data files are often compressed. Most often used is a Gzip compression, which results in an added *.gz at the end of the file name for example: example.fq.gz or example.fa.gz. When uploading files to galaxy, files get automatically unzipped, so this should not be an issue.
Importing and editing a published workflow
We have prepared a template workflow that you can edit and use in your genome assembly and annotation. The template workflow are split into different steps with several measures for checking the quality in between steps.
Let’s start by importing and exploring an initial QC workflow:
- In a new tab, open this link: https://usegalaxy.eu/u/matinnu/w/01rawqc
- On the top-right corner, click the
+button (import workflow) - Click
startusing this workflow - Click on the name of the newly imported workflow (
imported: 01_Raw_QC) and clickEdit - Explore the workflow and make changes if necessary
Discussion 02
- What is the input and final output of the workflow?
- What are the purpose of the two main tools in the workflow?
- Do you think Kraken2 is suited for long-read sequences?
Solution
TBD
When you are contempt with the workflow, click save. Next, we will run the analysis on barcode02.fastq (the smallest dataset).
- Click the “play” button on the top-right corner (
Run Workflow) - If the workflow does not show in detailed view, click
Expand to full workflow form - Set
Send results to a new historytoyes. - Change the history name to “01_Raw_QC_2021_sample02”.
- On the
1: input dataset, select2: barcode02.fastqas the input. - Click
Run Workflowon the top panel.
Working with Galaxy history
In Galaxy, each analysis will run on different history. You can have multiple history and switch between history depending on your needs.
We just run a simple QC to our raw reads, and it might take a while for the jobs to finish. Note that we have made some of the intermediate jobs hidden.
Discussion 03
When the job is done, let’s look at the output of the jobs in the history. Open the Nanoplot result by clicking the “eye” (
view data) icon.
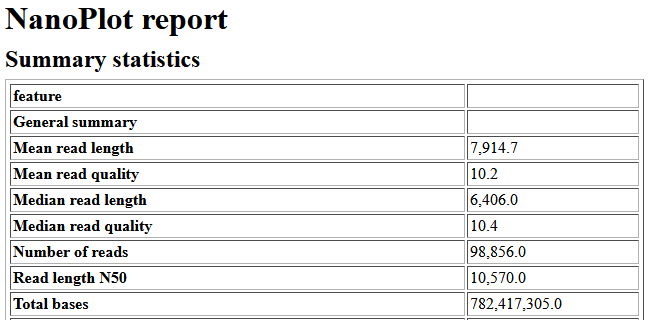
- Find out the value of these parameters:
- Total base
- N50
- Mean / Median Length
- Mean / Median Quality
- Your longest read
- Note that this sample have 506.4 MB (Mega Bytes) of data and a 262.4 MBp (Mega Base Pairs) of reads. Which one is more appropriate to measure the sequencing yield?
- What does N50 means? What is a good N50 value?
- There are 66% of reads that are above Q10. What does this mean? What is a Q-score?
- Do you think you have enough data for the assembly? What sequencing depth do you expect? How do you calculate a sequencing depth?
Solution
Example of Nanoplot result
Before the assembly a quality check is necessary. For that we can use Nanoplot with default settings. Nanoplot gives several output files, but you can get an easy overview with the HTML report. The most important numbers there the mean read quality and the read length N50. The read quality is denoted as a Phred score and is for nanopore reads typicall not very high. Anything above 7 should be fine. If you want to know more, it is possible to calculate back from the Phred score to the estimated probability of erroneous base pairs. The N50 is a measurement for the contiguity or the mean read/contig length. It is defined as the sequence length of the shortest contig at 50% of the total genome length. Low N50 (<1000bp) of the nanopore reads usually indicate sheared DNA, which can occur due to problems during the library prep or DNA extraction.
Let’s open the Krona Pie Chart to visualize the Kraken2 result.
- What does Kraken2 says about the majority of the reads? What species are assigned?
- Do you see potential contamination in your reads?
- Would you trust Kraken2 result as the taxonomic placement of your assembly or do you want to try another approach? Which database are being used by Krona for the taxonomic assignment?
Solution
TBD
Concluding the QC Analysis
Based on the QC, we have good quality and sufficient amount of reads for the assembly. Nevertheless, we might improve our assembly by filtering out the shorter reads (and perhaps bad quality reads?), which we will do in the next steps.
Key Points
You can start working in Galaxy by getting familiar to importing data, using history, and manipulating workflows.
Tools such as Nanoplot and Kraken2 can give you information about the quantity (yield), quality (Q-score, N50, and read distribution), and possible contamination.
QC are valuable to decide how the assembly process should be approached. It can tell us whether more sequencing data need to be generated or a certain part of the data need to be filtered. Overall, the decision might be a case by case basis.